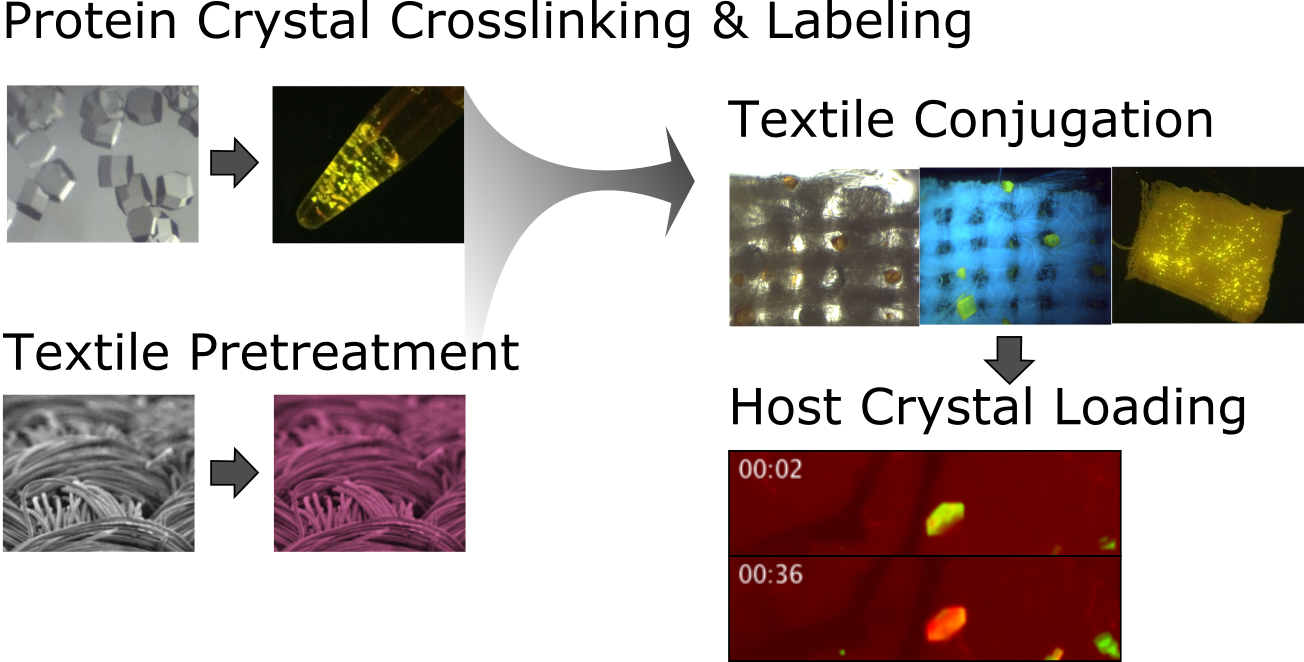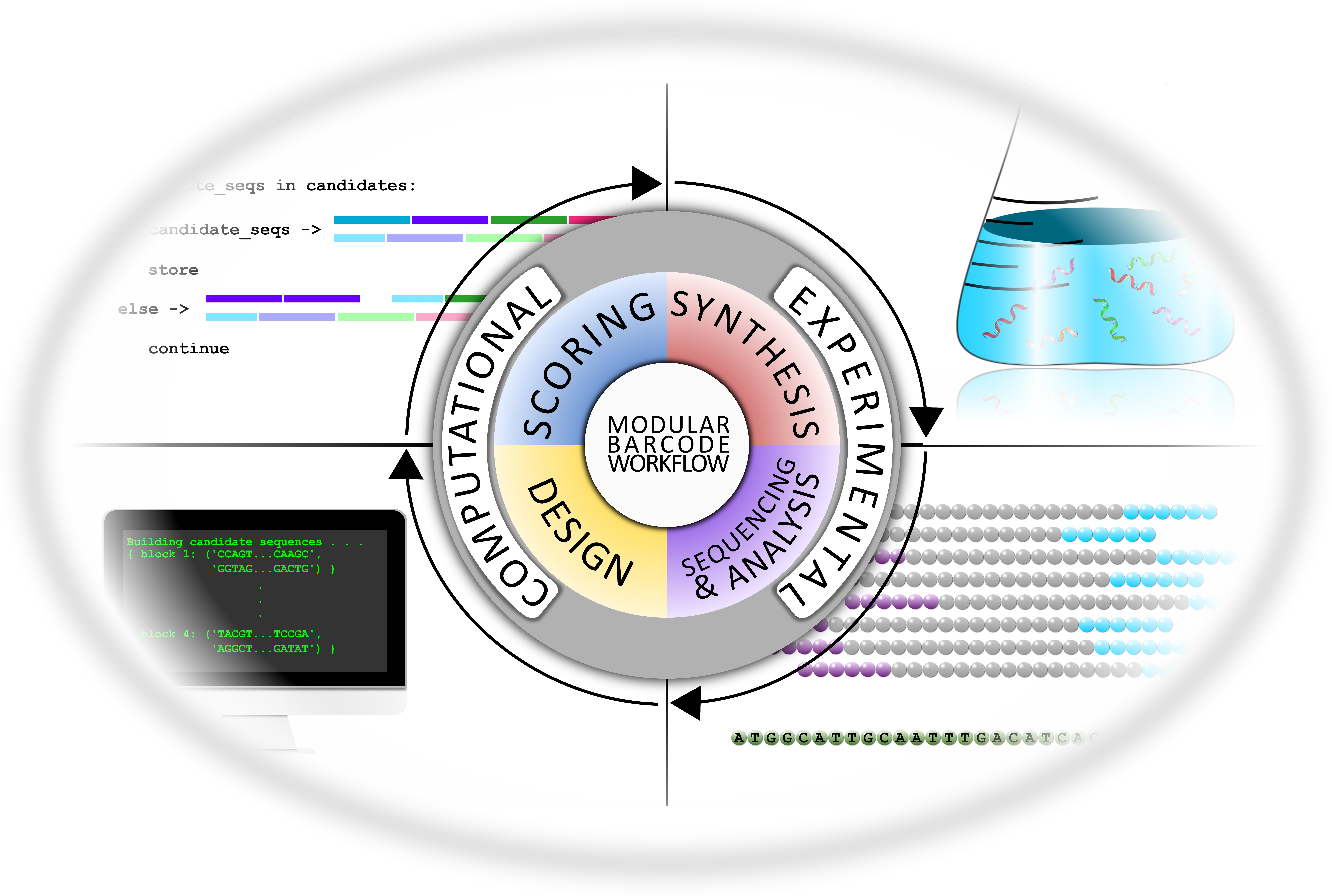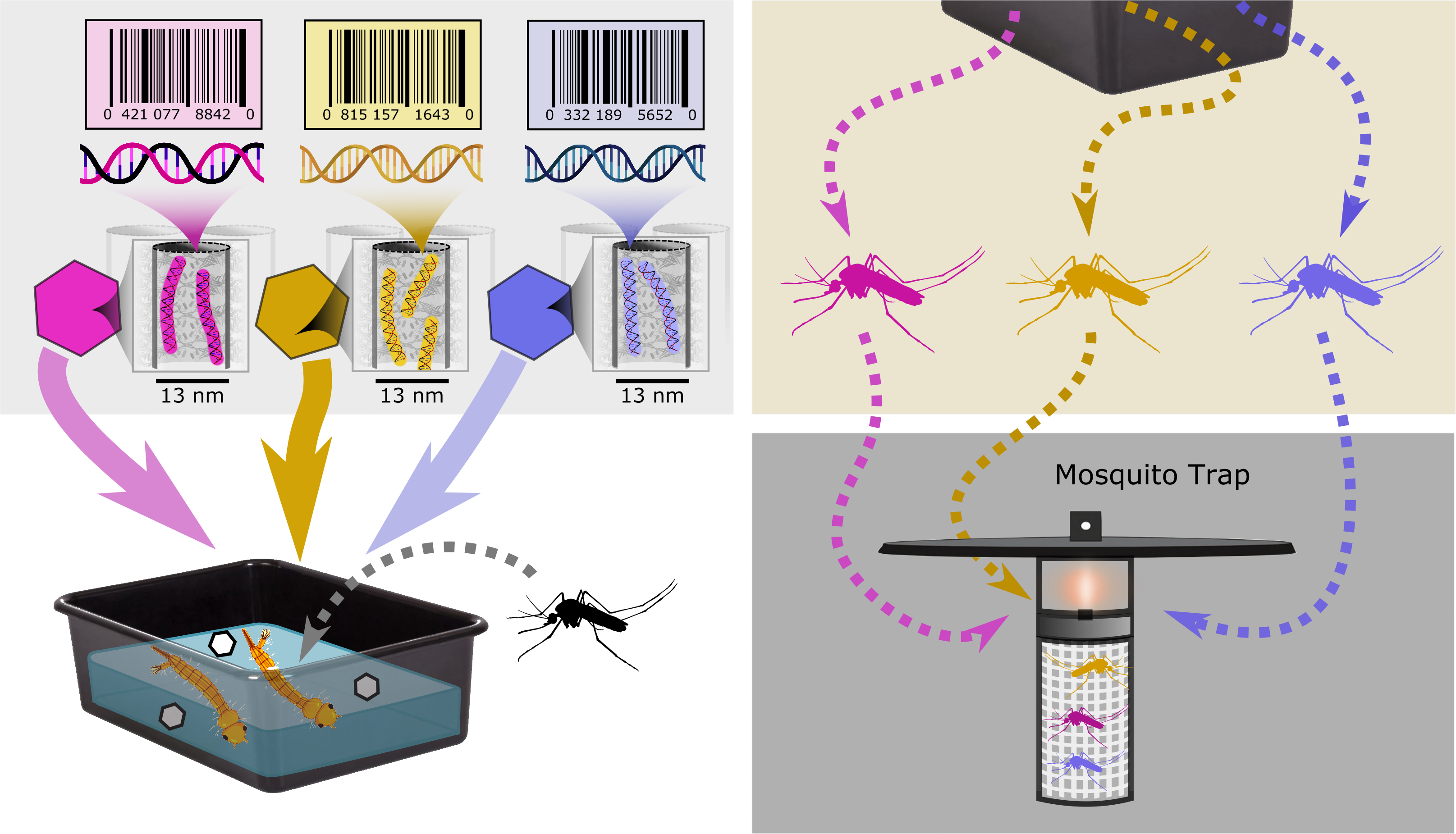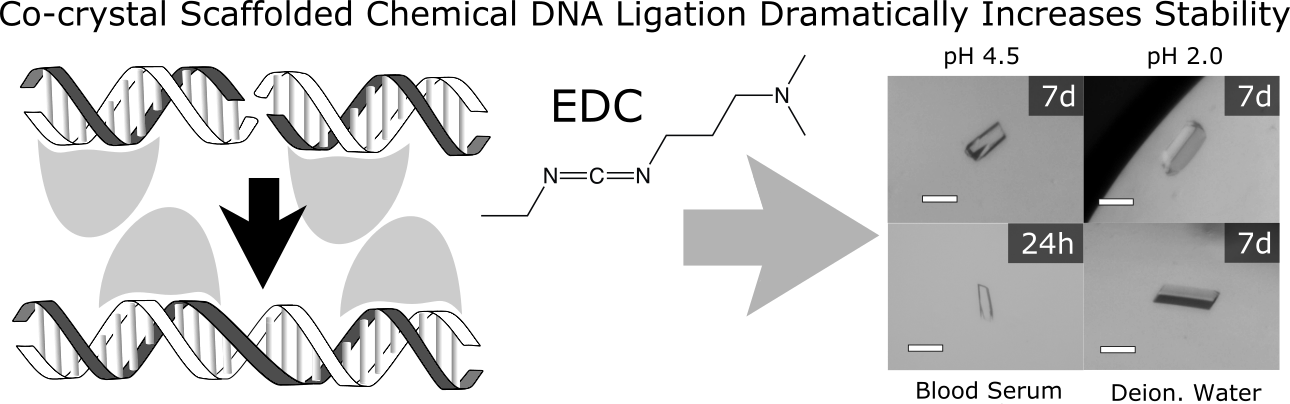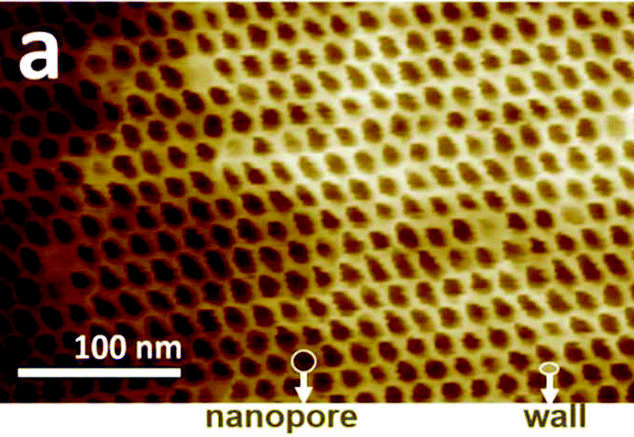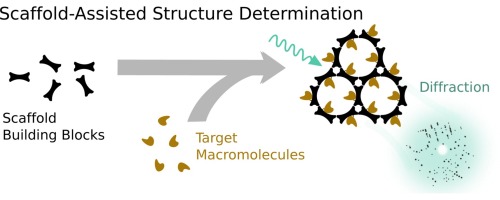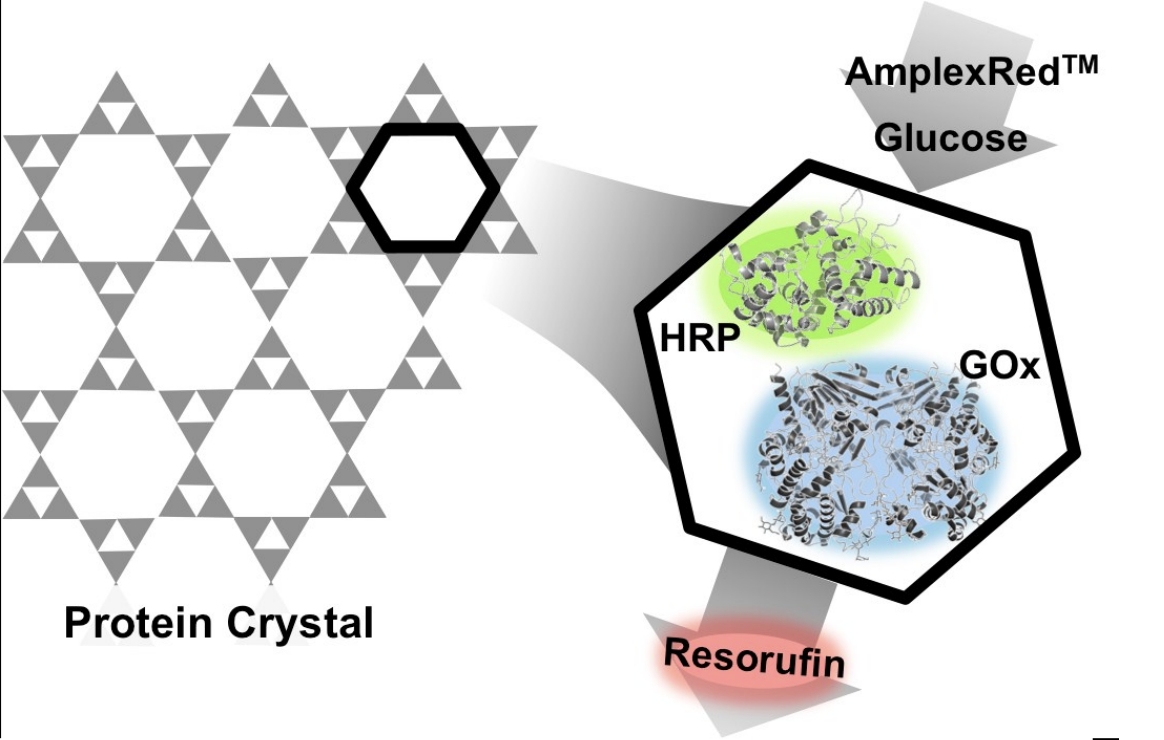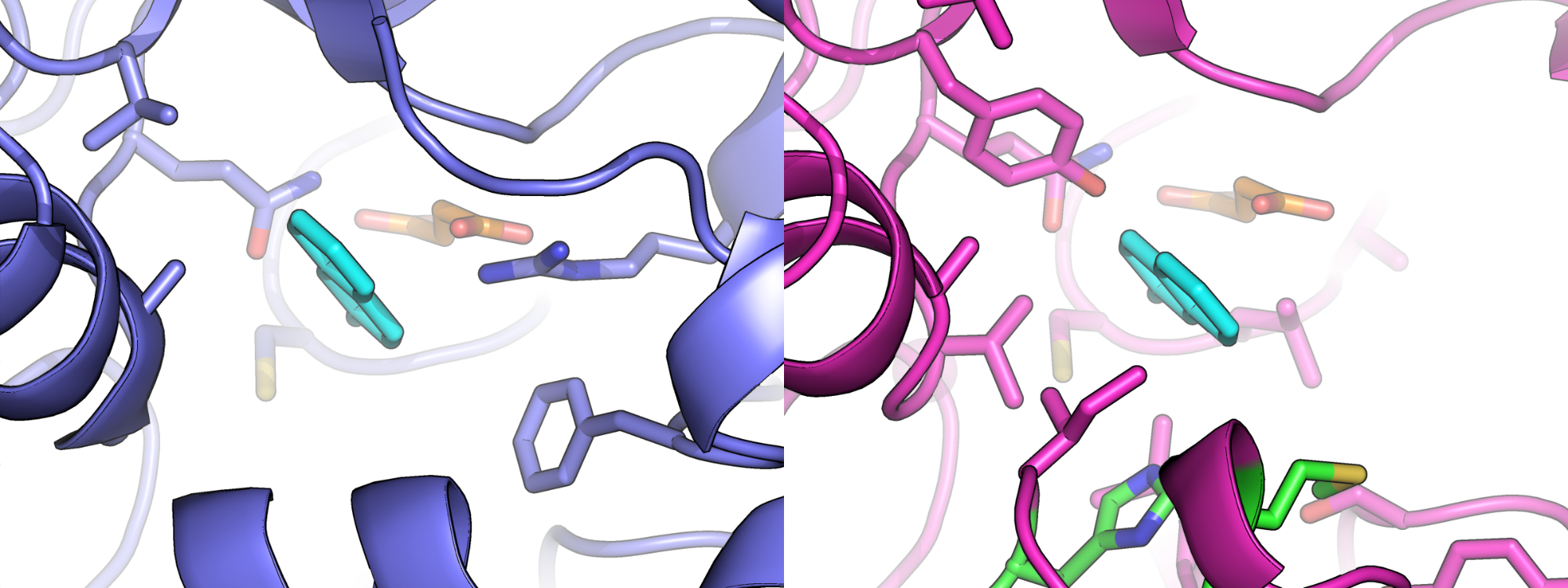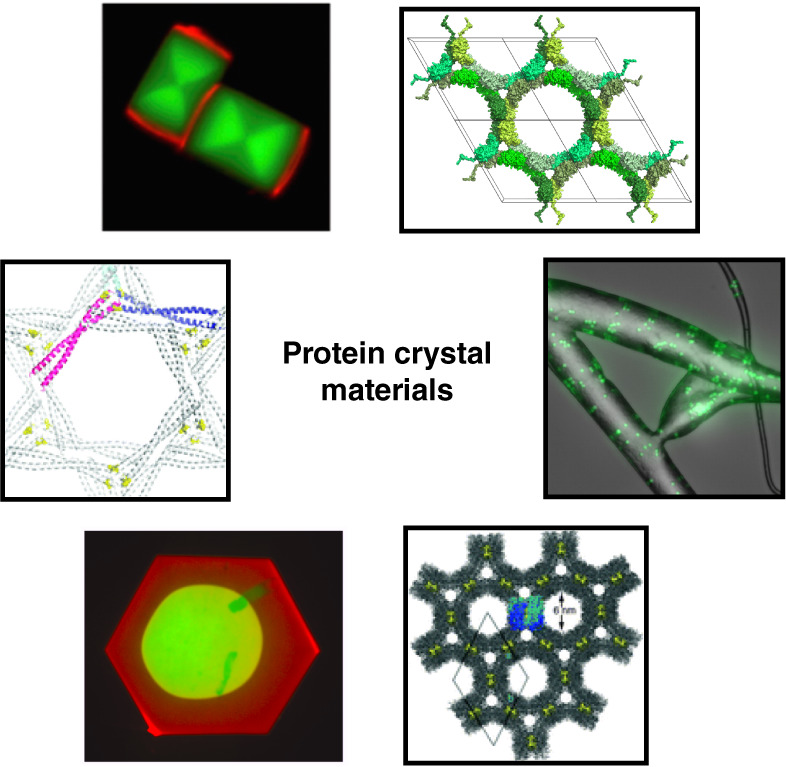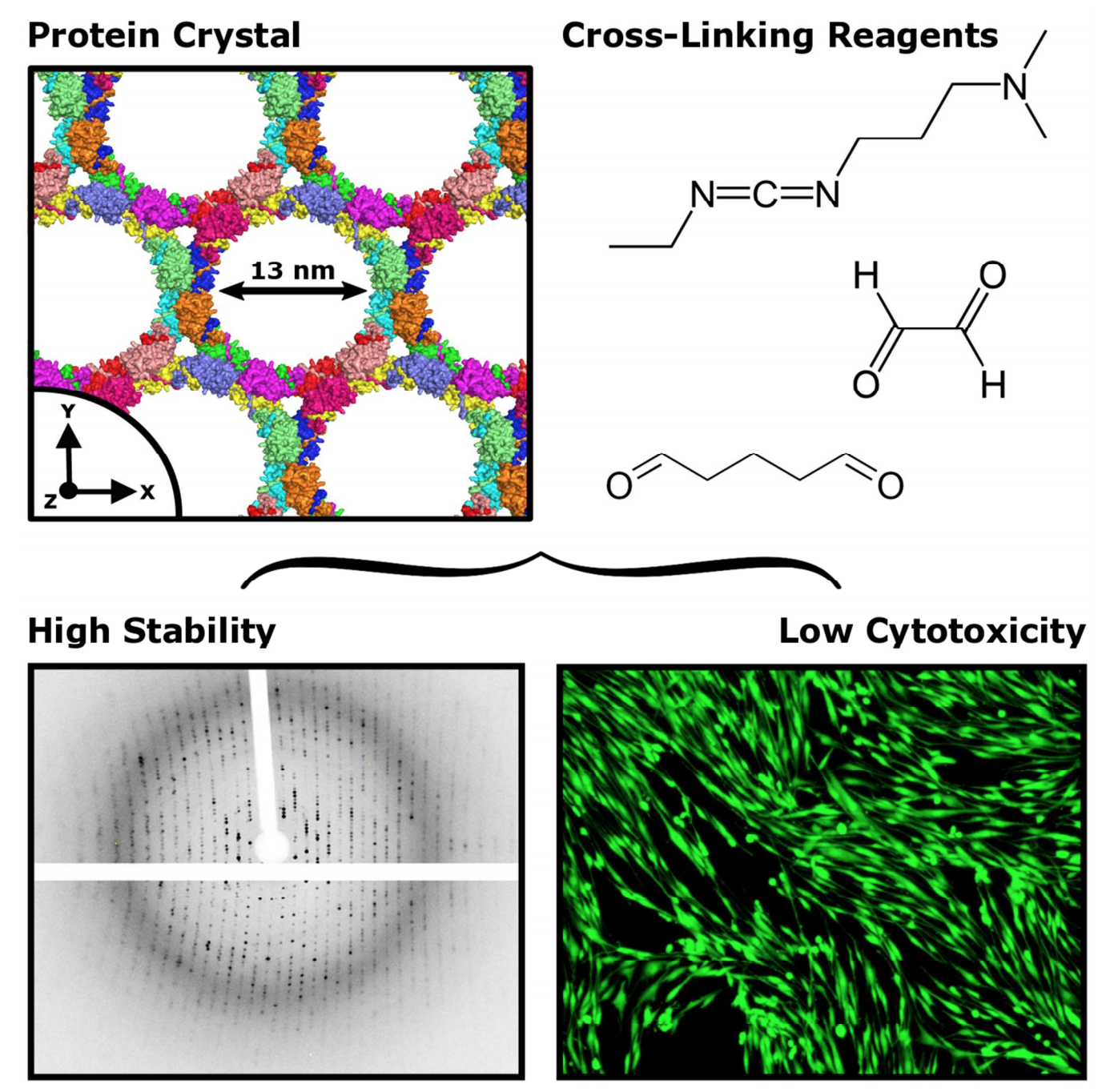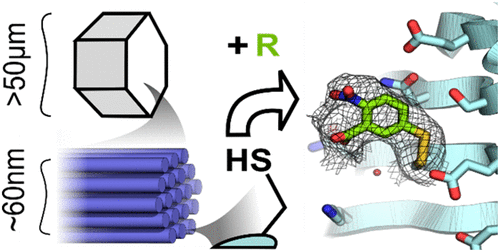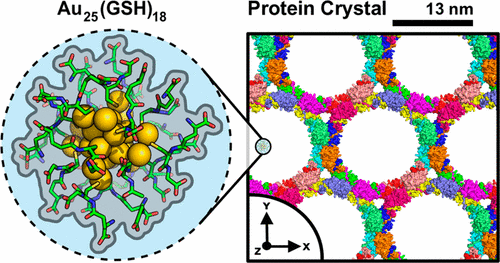
|
|
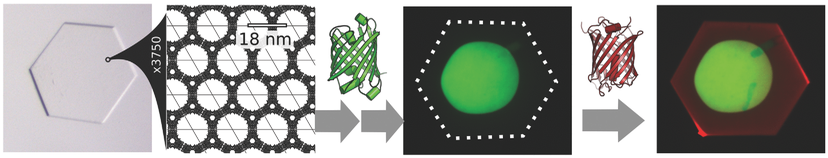
Non-bulk-like Solvent Behavior in the Ribosome Exit Tunnel
Del Lucent, Christopher Snow, Colin Aitken, Eric Sorin, Sung-Joo Lee & Vijay S. Pande.
PLoS Comp. Bio. (2010), 6, e1000963.
|
|
|
Side-chain recognition and gating in the ribosome exit tunnel
Paula Petrone, Christopher D. Snow, Del Lucent & Vijay S. Pande.
Proc. Natl. Acad. Sci. USA (2008) 105(43), 16549-16554.
|
|
|
Electric Fields at the Active Site of an Enzyme: Direct Comparison of Experiment with Theory
Ian Suydam, Christopher D. Snow, Vijay S. Pande & Stephen G. Boxer.
Science (2006)
|

|
|
Kinetic Definition of Protein Folding Transition State Ensembles and Reaction Coordinates
Christopher D. Snow, Young Min Rhee, & Vijay S. Pande.
Biophysical Journal (2006)
|
|
|
Direct calculation of the binding free energies of FKBP ligands
Hideaki Fujitani, Yoshiaki Tanida, Masakatsu Ito, Guha Jayachandran, Christopher D. Snow, Michael R. Shirts, Eric J. Sorin, & Vijay S. Pande.
Journal of Chemical Physics (2005) 123(8),084108
|
|
|
|
How well can simulation predict kinetics and thermodynamics of protein folding?
Christopher D. Snow, Eric Sorin, Young Min Rhee, & Vijay S. Pande.
Annual Review of Biophysics and Biomolecular Structure
|
|
|
Dimerization of the p53 oligomerization domain: Identification of a folding nucleus by molecular dynamics simulations
Lillian T. Chong, Christopher D. Snow, Young Min Rhee, & Vijay S. Pande.
Journal of Molecular Biology (2004) 345(4), 869-78.
|
|
|
Trp zipper folding kinetics
by molecular dynamics and temperature-jump spectroscopy
Christopher D. Snow, Linlin Qiu, Deguo Du, Feng Gai, Stephen J. Hagen, & Vijay S. Pande.
Proc. Natl. Acad. Sci. USA (2004) 101(12), 4077-4082.
|
|
|
|
Using path sampling to build better Markovian state models: Predicting the folding rate and mechanism of a tryptophan zipper beta hairpin
Nina Singhal, Christopher D. Snow, and Vijay S. Pande.
The Journal of Chemical Physics (2004) 121(1) 415-425.
|
|
|
Absolute comparison of simulated and experimental protein-folding dynamics
Christopher D. Snow, Houbi Nguyen, Vijay S. Pande, Martin Gruebele.
Nature AOP, published online 20 October 2002; doi:10.1038/nature01160.
Nature (2002) 420(6911), 102-106.
Up Front: Science Behind the Screens
HHMI Bulletin - M. Mitchell Waldrop
|
|
|
The Trp Cage: Folding Kinetics and Unfolded State
Topology via Molecular Dynamics Simulations
Christopher D. Snow, Bojan Zagrovic, Vijay S. Pande.
J. Am. Chem. Soc. (2002) 124(49), 14548-14549.
Native-like Mean Structure in the Unfolded Ensemble of Small Proteins
Bojan Zagrovic, Christopher D. Snow, Siraj Khaliq, Michael R. Shirts, Vijay S. Pande.
J. Mol. Biol. (2002) 323(1), 153-164.
|
|
|
Simulation of folding of a small alpha-helical
protein in atomistic detail using worldwide-distributed computing
Bojan Zagrovic, Christopher D. Snow, Michael R. Shirts, Vijay S. Pande.
J. Mol. Biol. (2002) 323(5), 927-937.
Atomistic protein folding simulations on the submillisecond time scale using worldwide distributed computing
Vijay S. Pande, Ian Baker, Jarrod Chapman, Sidney Elmer, Stefan M. Larson, Young Min Rhee, Michael R. Shirts,
Christopher D. Snow, Eric J. Sorin, Bojan Zagrovic.
Biopolymers (2002) 68(1): 91-109.
|
|
|
Folding@Home and Genome@Home: Using distributed
computing to tackle previously intractable problems in computational biology
Stefan M. Larson, Christopher D. Snow, Michael R. Shirts, Vijay S. Pande.
Computational Genomics, Richard Grant, editor, Horizon Press
|