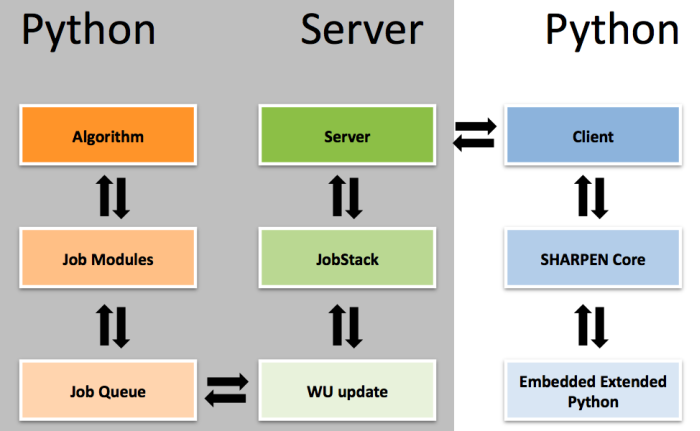
The first SHARPEN project will consist of careful tests of the new core -- we have developed an very different architecture. The platform is designed for rapid prototyping and testing of a variety of algorithms. Sidechain repacking, loop rebuilding, protein design, backbone search. As a rule, individual calculations do not require CPU-days. Instead, we bundle many small calculations into a full work unit. This is quite different from traditional Folding@Home calculations so we anticipate the need for careful beta testing.
Project: Cytochrome P450 structure prediction
Cytochrome P450s are large enzymes of great importance as models for protein chemistry and evolution and play key roles in human health. Their myriad biological functions include drug metabolism and steroid and secondary metabolite biosynthesis. More than 7,700 sequences are known. In humans, P450s are responsible for the disposition of many xenobiotics, including pharmaceuticals. Most drugs are metabolized in the liver by P450s during initial clearance.
Due to their medical importance, structure determination of new P450 enzymes is of high intrinsic interest. Unfortunately, mammalian cytochrome P450s have proven difficult to manipulate, making progress in understanding their structure and function slow. Only 6 structures of distinct human P450s have been published. Notably, CYP102A1, the soluble P450 from Bacillus megaterium (BM3), has been a heavily studied model system in place of the mammalian P450s. We are preparing to publish the first structure determination for a BM3 homolog, CYP102A2 from Bacillus subtilus.
Interest in accurate P450 structure prediction is also high. Many P450 homology models have been reported over the last 20 years. Natural P450 crystal structures share a core structure that may be aligned to ~3 Angstroms despite a low average pairwise sequence identity of 25%. However, to help explain enzyme stability and specificity, homology models must be refined to high resolution and validated by thorough comparison to experimental data. Accordingly we are developing new algorithms for structure refinement and also solving new P450 structures with X-ray crystallography.
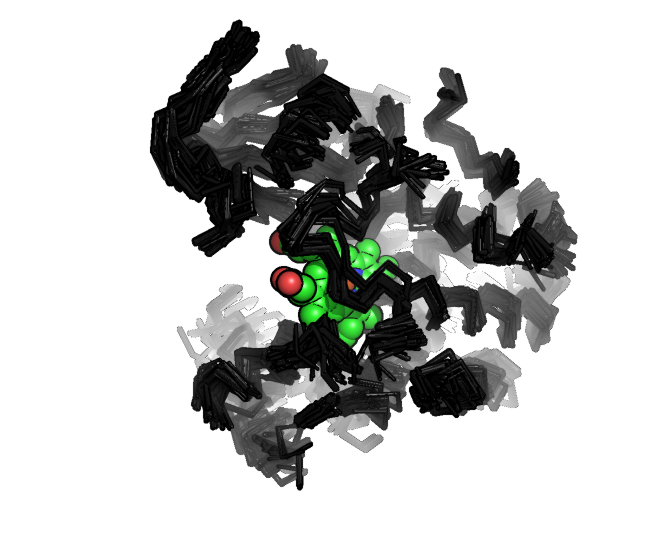
Project: Understanding P450 drug interactions
We are attempting to rationalize large datasets collected in the Arnold group that screen panels of substrates against a diverse panel of P450 enzymes. The best datasets include not just active/inactive enzyme patterns but include the distribution of products. We are thus attempting to build and train models for P450 regioselectivity. Such models could eventually be useful to predict drug toxicity, or to determine which synthetic enzyme would be the most productive catalyst for production of useful quantities of a drug that would otherwise be very diffeicult to synthesize.
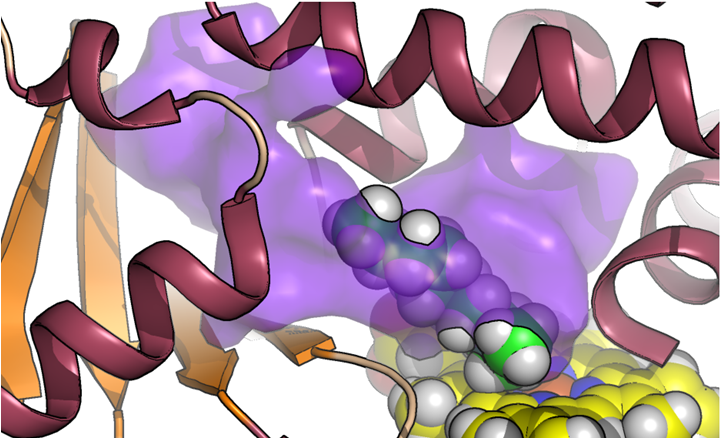
Project: Cellulase Library Design
The Arnold group is engineering new cellulases for biofuels. The project was recently described as one of the Technology Review 10 emerging technologies for 2008. In collaboration with DNA 2.0 we are using the power of recombination to engineer stabilized cellobiohydrolases. SHARPEN plays into this effort by allowing us to test new enzyme library design algorithms.
