PubMed Search |
|
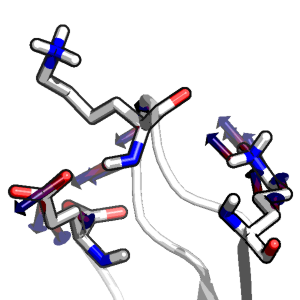
Polarizable Protein Packing Albert Ng and Christopher Snow Accepted, Journal of Computational Chemistry (2010) |
|
Combinatorial alanine substitution enables rapid optimization of cytochrome P450BM3 for selective hydroxylation of large substrates. Jared Lewis, Simone Mantovani, Yu Fu, Christopher Snow, Russell Komor, Chi-Huey Wong, and Frances Arnold. Chem. BioChem. (2010), 11, 2502-2505. |

|
Efficient screening of fungal cellobiohydrolase class I enzymes for thermostabilizing sequence blocks by SCHEMA structure-guided recombination Pete Heinzelman, Russell Komor, Arvind Kanaan, Philip Romero, Xinlin Yu, Shannon Mohler, Christopher Snow and Frances Arnold. Protein Engineering, Design and Selection (2010) dio: 10.1093/protein/gzq063 |
|
Non-bulk-like Solvent Behavior in the Ribosome Exit Tunnel. Del Lucent, Christopher Snow, Colin Aitken, Eric Sorin, Sung-Joo Lee & Vijay S. Pande. Accepted, PLoS Computational Biology. |

|
SCHEMA Recombination of a Fungal Cellulase Uncovers a Single Mutation that Contributes Markedly to Stability Pete Heinzelman, Christopher D. Snow, Matthew A. Smith, Xinlin Yu, Arbind Kannan, Kevin Boulware, Alan Villalobos, Sridhar Govindarajan, Jeremy Minshull, and Frances H. Arnold. J. Biol. Chem. (2009) 284:26229-26233. |
|
A Family of Thermostable Fungal Cellulases Created by Structure-Guided Recombination Pete Heinzelman, Christopher D. Snow, Indira Wu, Catherine Nguyen, Alan Villalobos, Sridhar Govindarajanm Jeremy Minshull and Frances H. Arnold. Proc. Natl. Acad. Sci. USA (2009) 106(14):5610-5615. |
|
SHARPEN: Systematic Hierarchical Algorithms for Rotamers and Proteins on an Extended Network Ilya V. Loksha, James R. Maiolo III, Cheng Hong, Albert Ng, and Christopher D. Snow. Journal of Computational Chemistry (2009) 30(6):999-1005. |

|
Side-chain recognition and gating in the ribosome exit tunnel. Paula Petrone, Christopher D. Snow, Del Lucent & Vijay S. Pande. Proc. Natl. Acad. Sci. USA (2008) 105(43), 16549-16554. |
|
Evolutionary history of the emergence of a specialized cytochrome P450 propane monooxygenase Rudi Fasan, Yergalem T. Meharenna, Christopher D. Snow, Thomas L. Poulos, and Frances H. Arnold. Journal of Molecular Biology (2008) 383(5), 1069-1080. |
|
Hunting for predictive computational drug discovery models Christopher D. Snow. Expert Review of Anti-infective Therapy (2008) 6(3) |
|
A diverse family of thermostable cytochrome P450s created by recombination of stabilizing fragments. Yougen Li, Alan D. Drummond, Andrew M. Sawayama, Christopher D. Snow, Jesse D. Bloom, Frances H. Arnold. Nature Biotechnology (2007) |
|
Electric Fields at the Active Site of an Enzyme: Direct Comparison of Experiment with Theory Ian Suydam, Christopher D. Snow, Vijay S. Pande & Stephen G. Boxer. Science (2006) |

|
Kinetic Definition of Protein Folding Transition State Ensembles and Reaction Coordinates. Christopher D. Snow, Young Min Rhee, & Vijay S. Pande. Biophysical Journal (2006) |
|
Direct calculation of the binding free energies of FKBP ligands. Hideaki Fujitani, Yoshiaki Tanida, Masakatsu Ito, Guha Jayachandran, Christopher D. Snow, Michael R. Shirts, Eric J. Sorin, & Vijay S. Pande. Journal of Chemical Physics (2005) 123(8),084108 |
|
How well can simulation predict kinetics and thermodynamics of protein folding?
Christopher D. Snow, Eric Sorin, Young Min Rhee, & Vijay S. Pande. Annual Review of Biophysics and Biomolecular Structure |
|
Dimerization of the p53 oligomerization domain: Identification of a folding nucleus by molecular dynamics simulations Lillian T. Chong, Christopher D. Snow, Young Min Rhee, & Vijay S. Pande. Journal of Molecular Biology (2004) 345(4), 869-78. |
|
Trp zipper folding kinetics by molecular dynamics and temperature-jump spectroscopy Christopher D. Snow, Linlin Qiu, Deguo Du, Feng Gai, Stephen J. Hagen, & Vijay S. Pande. Proc. Natl. Acad. Sci. USA (2004) 101(12), 4077-4082. |
|
Using path sampling to build better Markovian state models: Predicting the folding rate and mechanism of a tryptophan zipper beta hairpin
Nina Singhal, Christopher D. Snow, and Vijay S. Pande . The Journal of Chemical Physics (2004) 121(1) 415-425. |
|
Absolute comparison of simulated and experimental protein-folding dynamics Christopher D. Snow, Houbi Nguyen, Vijay S. Pande, Martin Gruebele. Nature AOP, published online 20 October 2002; doi:10.1038/nature01160. Nature (2002) 420(6911), 102-106. Up Front: Science Behind the Screens HHMI Bulletin - M. Mitchell Waldrop |
|
hSurface Salt Bridges, Double-Mutant Cycles, and Protein Stability: an Experimental and Computational Analysis of the Interaction of the Asp 23 Side Chain with the N-Terminus of the N-Terminal Domain of the Ribosomal Protein L9. Donna L. Luisi, Christopher D. Snow, Jo-Jin Lin, Zachary S. Hendsch, Bruce Tidor, Daniel P. Raleigh. Biochemistry (2003) 42(23), 7050-7060. |
|
The Trp Cage: Folding Kinetics and Unfolded State
Topology via Molecular Dynamics Simulations. Christopher D. Snow, Bojan Zagrovic, Vijay S. Pande. J. Am. Chem. Soc. (2002) 124(49), 14548-14549. Native-like Mean Structure in the Unfolded Ensemble of Small Proteins Bojan Zagrovic, Christopher D. Snow, Siraj Khaliq, Michael R. Shirts, Vijay S. Pande. J. Mol. Biol. (2002) 323(1), 153-164. |
|
Simulation of folding of a small alpha-helical
protein in atomistic detail using worldwide-distributed computing. Bojan Zagrovic, Christopher D. Snow, Michael R. Shirts, Vijay S. Pande. J. Mol. Biol. (2002) 323(5), 927-937. Atomistic protein folding simulations on the submillisecond time scale using worldwide distributed computing. Vijay S. Pande, Ian Baker, Jarrod Chapman, Sidney Elmer, Stefan M. Larson, Young Min Rhee, Michael R. Shirts, Christopher D. Snow, Eric J. Sorin, Bojan Zagrovic. Biopolymers (2002) 68(1): 91-109. |
|
Folding@Home and Genome@Home: Using distributed
computing to tackle previously intractable problems in computational biology. Stefan M. Larson, Christopher D. Snow, Michael R. Shirts, Vijay S. Pande. Computational Genomics, Richard Grant, editor, Horizon Press |
|
RNA binding to the aminoglycoside antibiotic Apramycin Spring 2002 Professor Puglisi, Stanford, Graduate Student Simulated Annealing Fragment Monte Carlo: Protein Structure Prediction Fall 2001 to Winter 2001 Professor Levitt, Stanford, Graduate Student Preparation for CASP5 Distributed Computing Protein Folding Spring 2001 to Summer 2001 Professor Pande, Stanford, Visiting Researcher Collaboration with Professor Gruebele at the University of Illinois 2002 Kollman Satellite Meeting: Biophysical Society Electrostatic Analysis of NTL9 Spring 2000 to Fall 2001 Professor Tidor, MIT, Undergraduate Researcher Collaboration with Professor Dan Raleigh at SUNY Stony Brook 2001 Pfizer Undergraduate Fellow Conference Electrostatic Ranking of Myoglobin Chimera Fall 1998 to Summer 1999 Professor Tidor, MIT, Undergraduate Researcher 1999 Protein Society Meeting |